GROMACS tools for analyzing membrane trajectories
Membrane undulation spectrumI wrote a gromacs tool to calculate the undulation spectrum from bilayer simulations. For the theory, I referred to the following papers:
I used the code to analyze simulations in my first fullerene paper, ref. [1] above; so if you use the tool, please to cite that paper. The code works with topologies from gromacs 3.x, so if you are using gromacs 4.x you can (a) adapt the code or (b) make a topology for gromacs 3. Please do let me know if you find mistakes in the code! |
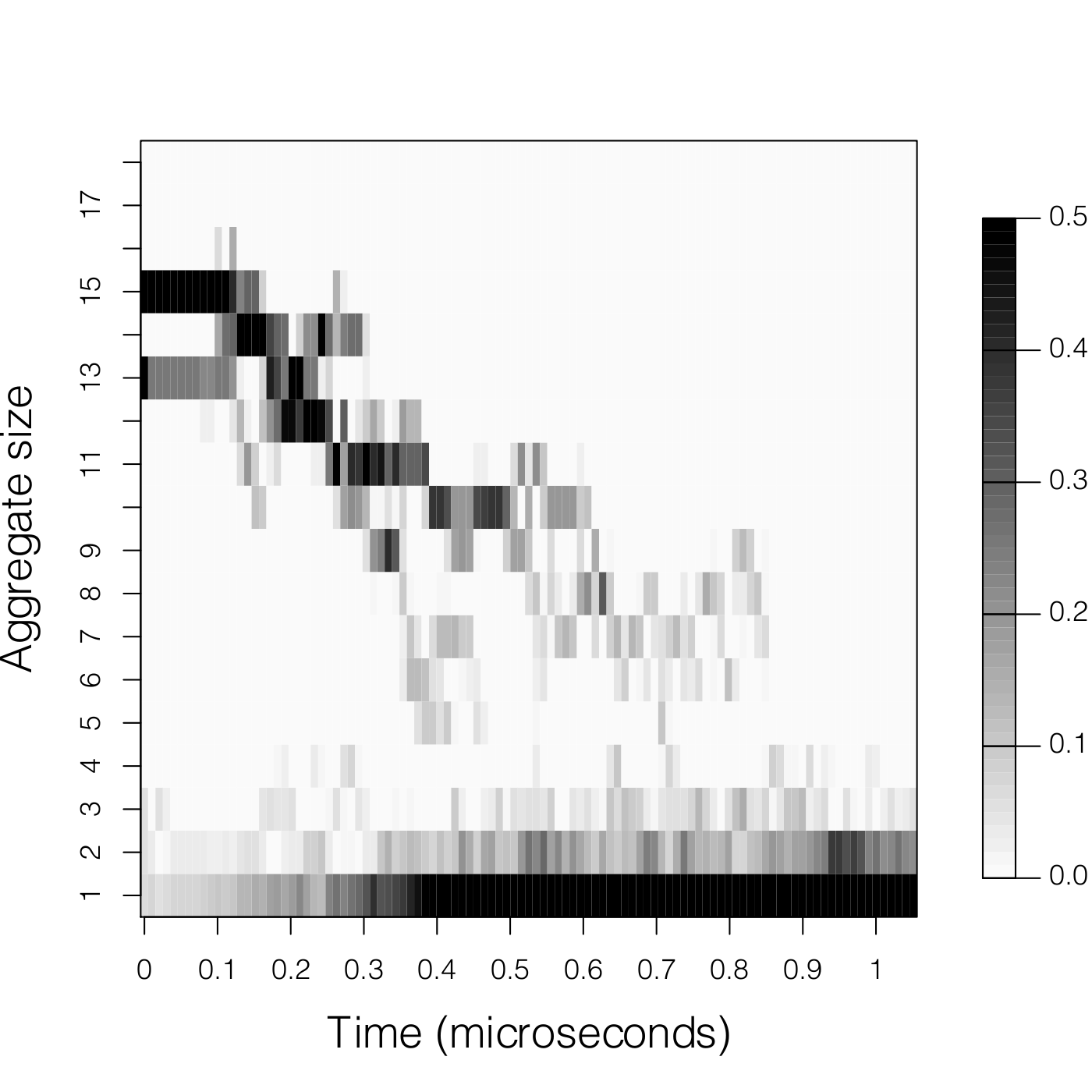
|
Aggregationg_aggregate.zip: click here to download.If you use this tool, please cite: Lipid Membranes as Solvents for Carbon Nanoparticles. J. Barnoud, G. Rossi, L. Monticelli, Phys. Rev Letters, (2014), 112, 068102 |
|
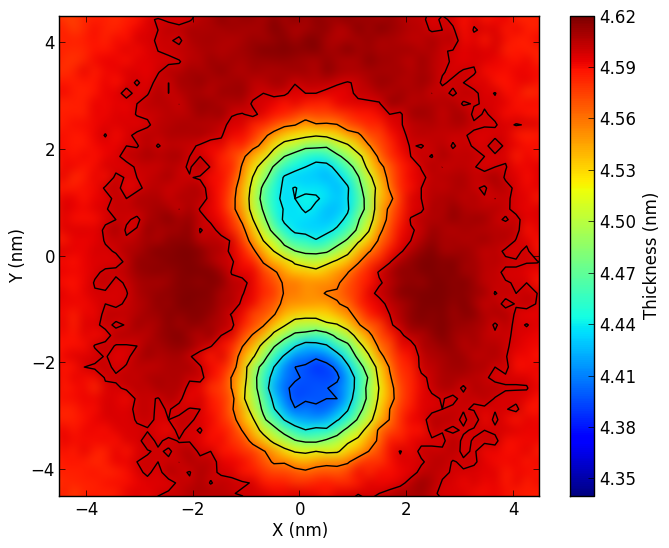
Membrane thicknessg_thickness.zip: click here to download.If you use this tool, please cite: Free Energy of WALP23 Dimer Association in DMPC, DPPC, and DOPC bilayers N. Castillo, L. Monticelli, Jonathan Barnoud, and D.P. Tieleman Chem. Phys. Lipids (2013), 169, 95-105. |
|
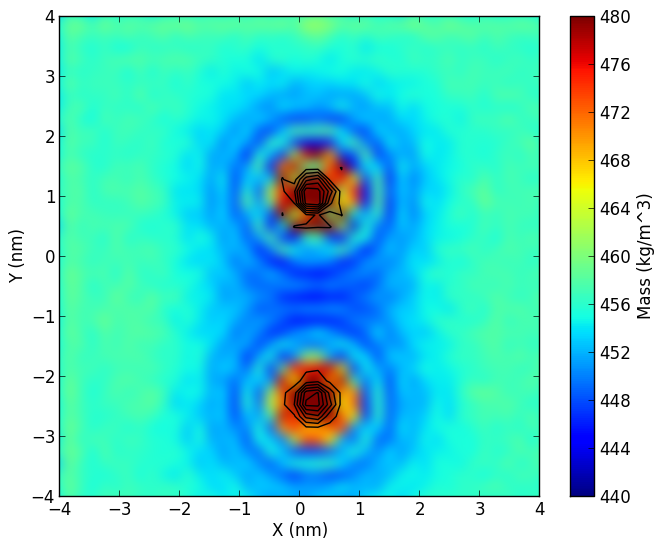
Membrane densityg_mydensity.zip: click here to download.If you use this tool, please cite: Free Energy of WALP23 Dimer Association in DMPC, DPPC, and DOPC bilayers N. Castillo, L. Monticelli, Jonathan Barnoud, and D.P. Tieleman Chem. Phys. Lipids (2013), 169, 95-105. |
|
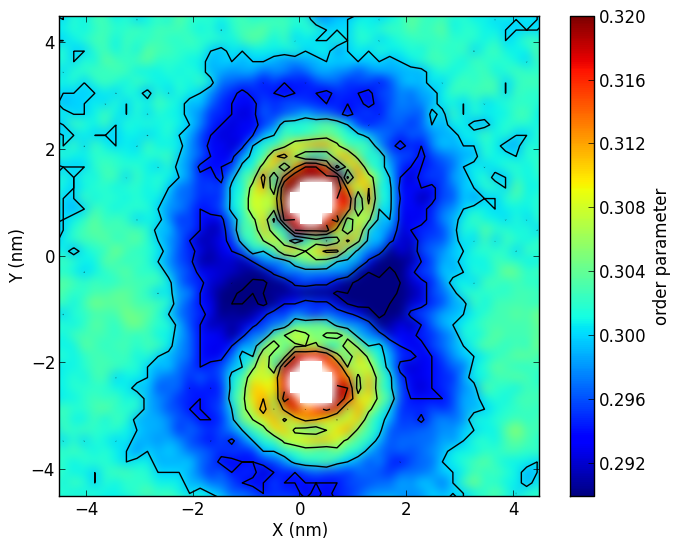
Membrane orderingg_ordercg.zip: click here to download.If you use this tool, please cite: Free Energy of WALP23 Dimer Association in DMPC, DPPC, and DOPC bilayers N. Castillo, L. Monticelli, Jonathan Barnoud, and D.P. Tieleman Chem. Phys. Lipids (2013), 169, 95-105. |
To make figures like the ones above:dispgrid.zip: click here to download.If you use this tool, please cite: Free Energy of WALP23 Dimer Association in DMPC, DPPC, and DOPC bilayers N. Castillo, L. Monticelli, Jonathan Barnoud, and D.P. Tieleman Chem. Phys. Lipids (2013), 169, 95-105. |